Salmonella Typhimurium employ spermidine to exert protection against ROS-
1
mediated cytotoxicity and rewires host polyamine metabolism to ameliorate its
2
survival in macrophages
3
Abhilash Vijay Nair
a
, Anmol Singh
a
, R. S. Rajamani
b
, Dipshikha Chakravortty
a,c
#
4
5
a
Department of Microbiology and Cell Biology, Division of Biological Sciences, Indian Institute
6
of Science, Bengaluru, India
7
b
Molecular Biophysiscs Unit, Indian Institute of Science, Bangalore, India
8
c
Adjunct Faculty, School of Biology, Indian Institute of Science Education and Research,
9
Thiruvananthapuram
10
11
Running Head: Salmonella mounts an antioxidative response using de-novo and host-acquired
12
spermidine
13
14
#Address correspondence to Dipshikha Chakravortty, [email protected], Tel: 0091 80 2293 2842,
15
Fax: 0091 80 2360 269
16
Keywords: Spermidine, Macrophages, Antioxidative response, Glutathionyl-spermidine
17
synthetase, D,L-α-difluoromethylornithine
18
19
20
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint

Abstract
21
Salmonella infection involves a cascade of attacks and defence measures. After breaching the
22
intestinal epithelial barrier, Salmonella is phagocytosed by the macrophages, inside which, the
23
bacteria face multiple stresses and, consequently, employ appropriate countermeasures. We show
24
that, in Salmonella, the polyamine spermidine activates a stress response mechanism by regulating
25
critical antioxidant genes. Salmonella Typhimurium mutants for spermidine transport and
26
synthesis cannot mount an antioxidative response, resulting in high intracellular ROS levels. These
27
mutants are also compromised in their ability to be phagocytosed by macrophages. Furthermore,
28
it regulates a novel enzyme in Salmonella, Glutathionyl-spermidine synthetase (GspSA), which is
29
known to prevent the oxidation of proteins in E.coli. Moreover, the spermidine mutants and the
30
GspSA mutant show significantly reduced survival in the presence of hydrogen peroxide in vitro,
31
and lesser organ burden in the mouse model of Salmonella infection. Conversely, in macrophages
32
isolated from gp91phox
-/-
mice, we observed a rescue in the attenuated fold proliferation previously
33
observed upon infection. Interestingly, Salmonella upregulates polyamine biosynthesis in the host
34
through its effectors from SPI-1 and SPI-2, which also solves the mystery of the attenuated
35
proliferation observed in spermidine transport mutants. Thus, inhibition of this pathway in the host
36
abrogates the proliferation of Salmonella Typhimurium in macrophages. From a therapeutic
37
perspective, inhibiting host polyamine biosynthesis using an FDA-approved chemopreventive
38
drug, D,L-α-difluoromethylornithine (DFMO), reduces Salmonella colonization and tissue
39
damage in the mouse model of infection, while enhancing the survival of infected mice. Therefore,
40
our work provides a mechanistic insight into the critical role of spermidine in stress resistance of
41
Salmonella. It also reveals a strategy of the bacteria in modulating host metabolism to promote
42
their intracellular survival and shows the potential of DFMO to curb Salmonella infection.
43
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Introduction
44
A condition with ample nutrients, and desired temperature, pH, oxygen concentration, and
45
osmolarity is usually considered optimal for the growth of microbes. An imbalance or any
46
alteration in these parameters impedes the growth and survival of the microbes, considered as
47
stress conditions. However, these parameters keep fluctuating in nature [1]. Hence, to persist and
48
survive in the natural environment, with unforeseen stressful or lethal conditions, they must be
49
adept in sensing, responding and adapting to them [2]. In the case of food-borne pathogens, they
50
face an array of various stresses in multiple environments in which they dwell, such as from the
51
natural habitat to commercial settings and inside the host system [3-5]. Salmonella is a food-borne
52
pathogen that enters the host system through contaminated food and water. In the host system, as
53
it traverses to the intestine, Salmonella faces multiple stressful conditions such as low pH, nutrient
54
deprivation, bile salt stress, competition with the resident microbes of the gut and
55
immunoglobulins, etc . Once it breaches the epithelial barrier, it is taken up by the macrophages at
56
the lamina propria, through which it disseminates throughout the host system. Macrophages are
57
phagocytic immune cells where Salmonella encounters a very hostile environment. Entry of the
58
pathogen into the cell cytoplasm leads to a burst of reactive oxygen species (ROS) and reactive
59
nitrogen species (RNS) [6, 7]. In macrophages, Salmonella resides in a specialised niche called
60
the Salmonella-containing vacuole (SCV), which presents multiple other stresses to the bacteria,
61
such as acidification, nutrient limitation and attack by the antimicrobial peptides. However,
62
Salmonella employs numerous weapons from its arsenal to counteract the stresses it faces within
63
the host macrophages.
64
Polyamines are a group of primordial stress response molecules in prokaryotes and eukaryotes [8].
65
Multiple research groups have elucidated the link between polyamines and bacterial stress
66
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
response. In Shigella spp. the polyamine spermidine accumulates during its infection into
67
macrophages, which increases the expression of katG and helps in bacterial antioxidant response
68
[9]. Moreover, spermidine localises to the surface of Pseudomonas aeruginosa and bind to the
69
lipopolysaccharide (LPS) to protect the cells from oxidative damage [10]. In the Gram-positive
70
bacteria, Streptococcus pyogenes, extracellular spermidine enhance the survival of the bacteria by
71
upregulating oxidative response genes [11]. A research group has shown that polyamines are vital
72
in resistance against nitrosative stress in Salmonella Typhimurium. Further, the group showed that
73
spermidine is required for the systemic infection of Salmonella Typhimurium in mice [12]. Thus,
74
it is conceivable that Salmonella Typhimurium utilises polyamines, such as spermidine, as a stress
75
response molecule; however, the mechanism remains elusive.
76
Here, we show that spermidine biosynthesis and transport mutants of Salmonella Typhimurium
77
exhibit reduced survival upon infection in RAW264.7 cells. This diminished proliferation is also
78
observed in mice models of Salmonella infection, which is rescued in gp91phox
-/-
mice. We
79
demonstrate that spermidine orchestrates the various arms of antioxidative response and aids in
80
tightly regulating intracellular ROS levels. We further identify a novel antioxidative enzyme, Gsp,
81
in Salmonella Typhimurium, which is regulated by spermidine. The fascinating question that arises
82
is why the transporter mutant shows reduced survival. To this, we find that Salmonella
83
Typhimurium harnesses the host polyamine biosynthesis for its survival. Furthermore, for the first
84
time, we show that an FDA-approved chemopreventive and anti-African Human Trypanosomiasis
85
drug that inhibit the polyamine biosynthesis in the host, is able to curb Salmonella infection in
86
mice models.
87
Material and methods
88
Bacterial strains and growth condition
89
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Salmonella enterica serovars Typhimurium (STM WT) wild type strain ATCC 14028s was used in
90
all experiments which was a kind gift from Prof. Michael Hensel, Abteilung Mikrobiologie,
91
Universität Osnabrück, 273 Osnabrück, Germany. The bacterial strain was cultured in Luria broth
92
(LB-Himedia) with constant shaking (175rpm) at 37°C orbital-shaker. Kanamycin,
93
Chloramphenicol and Ampicillin antibiotics (Kanamycin-50μg/ml, Cholramphenicol-20μg/ml and
94
Ampicillin-50μg/ml) were used wherever required. Strains were transformed with pFPV-m-cherry
95
plasmid for immunofluorescence assays. (Bacterial strain list in Supplementary table S-Table 1)
96
Bacterial gene knock-out and strain generation
97
The generation of gene knock-out in bacteria was done using the One-step chromosomal gene
98
inactivation protocol [13]. Briefly, the Kanamycin resistance gene and the Chloramphenicol
99
resistance gene amplified products were purified using chloroform-ethanol precipitation. Followed
100
by electroporation into the STM WT cells (expressing PKD-46 plasmid which provides the λ-Red
101
recombinase system) by a single pulse of 2.25 kV separately for the Kan
R
and Chlm
R
. The
102
transformant colonies were selected and patched on fresh plates and confirmed for knock-out using
103
PCR with primers designed corresponding to the ~100bp upstream and downstream of the genes
104
(knocked out) for the knock-out strains to observe a difference in PCR product size upon STM
105
ΔpotCD and STM ΔspeED knockout generation. For the generation of the double knock-out strain
106
(STM ΔpotCDΔspeED), the STM ΔpotCD (resistant to Kanamycin) was first transformed with the
107
plasmid pKD46 which provides the λ-Red recombinase system. To this transformed strain, the
108
purified PCR product to knock-out speED was electroporated to generate the STM
109
ΔpotCDΔspeED (resistant to Kanamycin and Chloramphenicol). For the generation of STM Δgsp,
110
Kanamycin resistance gene was amplified from pKD4 plasmid, and a similar protocol was used,
111
followed by selection on Kanamycin containing LB agar plates. For the generation of double gsp
112
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
spermidine mutants, STM Δgsp was electroporated with purified PCR product to knock-out speED
113
and potCD (both Chloramphenicol resistance cassette). (Knockout generation Primer list in
114
Supplementary table S-Table 2)
115
Cell culture and maintenance
116
RAW264.7 cells (murine macrophage cell line) were cultured in DMEM - Dulbecco’s Modified
117
Eagle Medium (Lonza) supplemented with 10% FBS (Gibco) and 1% Penicillin-streptomycin
118
(Sigma- Aldrich) at 37°C humidified chamber (Panasonic) with 5% CO
2
. For each experiment, the
119
cells were seeded onto the appropriate treated cell culture well plate at a confluency of 80% for
120
intracellular survival assay, and expression studies.
121
Gentamicin protection assay
122
The cells were infected with STM WT, STM ΔpotCD, STM ΔspeED, STM ΔpotCDΔspeED, STM
123
Δgsp and STM ΔkatG at MOI of 10 (for intracellular survival assay) and MOI 25 (for qRT-PCR).
124
After infecting the cell line with STM WT and the mutants, the plate was centrifuged at 700-900
125
rpm for 10 minutes to facilitate the proper adhesion. The plate was then incubated for 25 minutes
126
at 37°C humidified chamber and 5% CO
2
. Then the media was removed from the wells and washed
127
with 1X PBS. Fresh media containing 100 µg/mL gentamicin was added and again incubated for
128
60 minutes at 37°C and 5% CO
2
. The media was then removed, cells were washed with 1X PBS
129
twice, and fresh media containing 25µg/mL gentamicin was added. The plate was incubated at
130
37°C and 5% CO
2
till the appropriate time. For the intracellular survival assay, two time points
131
were considered 2 hours and 16 hours, and for qRT-PCR three time points were considered 2
132
hours, 6 hours and 16 hours. For phagocytosis assay upon opsonisation, all the strains were washed
133
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
with 1XPBS and incubated at 37°C with mouse complement sera for 1 hour before gentamicin
134
protection assay
135
Intracellular survival assay and phagocytosis assay
136
At the appropriate time post-infection, the cells were lysed using 0.1% Triton X followed by
137
addition of more 1X PBS and samples were collected. The collected samples were plated at the
138
required dilutions on LB agar plates and kept at 37°C. 12 hours post incubation the Colony forming
139
units (CFU) were enumerated for each plate.
140
The fold proliferation and invasion were determined as follows
141
Fold Proliferation = (CFU at 16 hours post-infection)/(CFU at 2 hours post-infection)
142
Percentage Phagocytosis = [(CFU at 2 hours post-infection)/(CFU of the Pre-inoculum)]×100
143
RNA isolation and qRT-PCR
144
RNA isolation was performed from infected cells after appropriate hours of infection with STM
145
WT, STM ΔpotCD, STM ΔspeED by RNA isolation was performed using TRIzol (Takara) reagent
146
according to manufactures’ protocol RNA was quantified using Thermo-fischer scientific Nano
147
Drop followed by running on 2% agarose gel for checking the quality. For cDNA synthesis, first
148
DNase I treatment with 3μg of isolated RNA was done at 37℃ for 60 minutes, which was then
149
stopped by heating at 65℃ for 10 minutes. Then RNA (free of DNA) was subjected to Reverse
150
transcription using Random hexamer, 5X RT buffer, RT enzyme, dNTPs and DEPC treated water
151
at 37°C for 15 minutes, followed by heating at 85℃ for 15 seconds. Quantitative real-time PCR
152
was done using SYBR green RT-PCR kit in BioRad qRT-PCR system. A 384 well plate with three
153
replicates for each sample was used. The expression levels of the gene of interest were measured
154
using specific RT primers. Gene expression levels were normalised to 16SrDNA primers of S.
155
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Typhimurium. Gene expression levels of eukaryotic gene of interest were normalised to beta-actin
156
of mouse/human as required. For expression studies in bacteria grown in LB media, the bacterial
157
samples were harvested at 3 hours, 6 hours, 9 hours and 12 hours post subculture in fresh LB media
158
in 1:100 ratio and 1mM H
2
O
2
was added to the broth, to study the gene expression in the presence
159
of oxidative stress. Then similar protocol was used to isolate total RNA using TRIzol (Takara)
160
reagent according to manufactures’ protocol (Expression Primer list in Supplementary table S-
161
Table 2)
162
Primary macrophages isolation and infection
163
Primary macrophages were isolated from C57BL/6 mice (male, 5-6 weeks old). The mice were
164
intraperitoneally injected with 8% Brewer’s Thioglycolate (HiMedia). Five days post injection the
165
primary macrophages were aseptically isolated by injecting ice-cold 1xPBS into the peritoneal
166
cavity the peritoneal lavage was collected. Any residual erythrocytes were lysed using RBC lysis
167
buffer ( Sigma- R7757) , and the isolated cells were maintained in complete RPMI 1640 media for
168
further experiments.
169
Intracellular Reactive oxygen species determination using H
2
DCFDA staining
170
Overnight cultures were sub-cultured in fresh LB media. Once the cultures reached OD 0.1 then
171
10
8
CFU/ml of each strain was incubated with 10µM of 2',7'-dichlorodihydrofluorescein diacetate
172
(H2DCFDA) (Sigma) in 1xPBS (pH 7.2) at 37°C for 30 minutes. The bacterial cells were
173
centrifuged and the cells were resuspended in 1xPBS (pH 7.2) with Hydrogen peroxide of different
174
concentrations (0mM- 10mM) , and incubated 37°C (orbital shaker) for 2 hours. The samples were
175
transferred to a 96 well ELISA plate and fluorescence was determined in Tecan-ELISA plate reader
176
Infinite series 200 ( Ex- 490nm/ Em- 520nm).
177
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Intracellular redox status determination
178
The STM WT, STM ΔpotCD, STM ΔspeED and STM Δgsp were transformed with pQE60-Grx1-
179
roGFP2 (a gift from Prof. Amit Singh, CIDR, IISc). Overnight cultures were sub-cultured in fresh
180
LB media. Once the cultures reached OD 0.1 then 10
8
CFU/ml of each strain was incubated with
181
Hydrogen peroxide (0mM-5mM) of different concentrations in 1xPBS (pH 7.2) with, and
182
incubated 37°C (orbital shaker) for 2 hours. The samples were centrifuged and resuspended in
183
fresh 1xPBS (pH 7.2). And tubes were analysed for GFP fluorescence at 408nm and 488nm
184
respectively using BD-FACS Verse flow cytometer (total 10,000 events for each sample). For
185
determination of intracellular redox status upon infection into RAW264.7 cells, 10
5
RAW264.7
186
cells seeded in a 24 well tissue culture plate, were infected with each of the strains harboring the
187
roGFP2 plasmid using gentamicin protection assay. After 16 hours post infection, the macrophages
188
were washed with 1X PBS and scrapped off using cell scraper and analysed for GFP fluorescence
189
at 408nm and 488nm respectively using BD-FACS Verse flow cytometer (total 10,000 events for
190
each sample).
191
In vitro sensitivity assays
192
Overnight cultures were sub-cultured in fresh LB media. Once the cultures reached OD 0.1 then
193
10
8
CFU/ml of each strain was incubated with Hydrogen peroxide or sodium nitrite of different
194
concentrations (0mM- 10mM) in 1xPBS(pH 7.2) (1xPBS of pH 5.4 was used for nitrite sensitivity
195
assay) with, and incubated 37°C (orbital shaker) for 2 hours. The samples were plated on SS agar
196
to enumerate the CFU and the percentage survival was determined as:
197
Percentage Survival : [ [CFU/ml for treated with H
2
O
2
/ NaNO
2
]/ [CFU/ml for untreated] ]×100
198
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Resasurin assay was used to determine the viable cells was performed in 96 well plate (triplicate
199
for each sample and concentration. Briefly, after incubation for 2 hours as previously performed,
200
Resasurin (0.2mg/ml in 1X PBS) was added (1:10 ratio) to each well of 96 well plate. The plate
201
was incubated at 37°C shaker incubator for 2 hours. The fluorescence was measure using Tecan
202
plate reader infinite series 200, with Ex-520nm and Em- 590nm. The values obtained as Relative
203
fluorescence units (RFU) and the percentage survival was determined as
204
Percentage survival = [RFU of sample with added hydrogen peroxide]/ [RFU of sample without
205
of hydrogen peroxide]
206
Immunoblotting
207
The bacterial strains were grown in LB media with added 1mM hydrogen peroxide until the log
208
phase of growth. The cells were centrifuged to remove the media, and the cells were resuspended
209
in the lysis buffer (Sodium chloride, Tris, EDTA, 10% protease inhibitor cocktail) after washing
210
with 1XPBS. The cells were lysed using sonication and centrifuged at 4°C to collect the cell lysate,
211
followed by estimation of total protein using the Bradford protein estimation method. 50µg of
212
protein was loaded onto a Polyacrylamide Gel Electrophoresis (PAGE) without β-mercaptoethanol
213
(prevent di-sulphde bond breakage as glutathionyl-spermidine modifies Cysteine residues through
214
a disulphide bond), then transferred onto 0.45μm PVDF membrane (GE Healthcare). 5% skimmed
215
milk (Hi-Media) in TTBS was used to block for 1hour at room temperature and then probed with
216
Anti-Spermidine primary (Novus Biologicals) and the secondary HRP-conjugated antibodies.
217
ECL (Biorad) was used for developing the blot, and images were captured using Chemi-
218
Doc(Biorad). All densitometric analysis was performed using the Image J. The normalisation was
219
done with respect to Ponceau S stained blot.
220
Transfection for knockdown
221
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
RAW 264.7 cells were seeded at a confluency of 50-60% 12 hours prior to transfecting using either
222
PEI (1:2 -DNA: PEI). We used two different constructs for knock-down of Odc1, A7 and F8 and
223
both as a mixed construct (A7:A8 at 1:1 ratio) and similarly for Srm , E9 and F1 and both as a
224
mixed construct (E9:F1 at 1:1 ratio) from the Sigma Mission shRNA library. 400ng of plasmid
225
DNA/well (ratio 260/280 ~1.8- 1.9) was used for transfection in a 24well plate. Cells were then
226
incubated for 8 hours at 37℃ in a humidified incubator with 5% CO2; after that, the media
227
containing transfecting DNA and reagents were removed, and cells were further incubated for 48
228
hours in complete media DMEM +10% FBS. Cells were harvested for further analysis or infected
229
with the required MOI using the gentamicin protection assay. (shRNA sequence list in
230
Supplementary table S-Table 3)
231
Immunofluorescence
232
After the appropriate incubation time after gentamicin protection assay, the media was removed,
233
and the cells were washed with 1X PBS and fixed with 3.5% Paraformaldehyde for 10 minutes.
234
The cells were then washed with 1X PBS, followed by incubation with the required primary
235
antibodies (anti-mouseLAMP1 and anti-Spermidine) in a buffer containing 0.01% saponin and 2%
236
BSA, and incubated at room temperature for 45-60 minutes. After washing with 1X PBS, the
237
secondary antibody tagged to a fluorochrome was added and incubated (anti-rat-Alexafluor488 for
238
LAMP1, anti-rabbit-Alexafluor647 for spermidine). The coverslips were then washed with PBS
239
and mounted on a clean glass slide using mounting media containing an anti-fade reagent and
240
observed under the confocal microscope (Zeiss 710 microscope, at 63X oil immersion, 2x319 3x
241
zoom, and 100X zoom for studying only bacterial samples, Zeiss 880 microscope, at 63X oil
242
immersion, 2x319 3x zoom).
243
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
The histopathological sections were deparaffinized and then incubated with the required primary
244
antibody (anti- Salmonella LPS) in a buffer containing 0.01% saponin and 2% BSA and incubated
245
at room temperature for 45-60 minutes. The primary antibody was then removed by washing with
246
1X PBS and then incubated with the appropriate secondary antibody tagged to a fluorochrome.
247
The sections were then washed with PBS and cover with coverslip after using mounting media
248
containing an anti-fade reagent. The coverslips were sealed with clear nail polish and observed
249
under the confocal microscope. For studying histopathology samples Zeiss 880 microscope 40X
250
oil immersion 2x319 3x zoom was used.
251
Intracellular RNS determination
252
For determination of intracellular RNS, a cell permeable nitric-oxide probe, 4, 5-
253
diaminofluorescein diacetate (DAF
2
A) (Sigma) was used. The protocol has been followed as Roy
254
Chowdhury A et. al. [14]. Briefly, After 16 hours of infection of RAW264.7 cells (transiently
255
knock down of Odc1) at MOI 10 with STM WT, the cells were incubated with fresh DMEM
256
containing 5µM of DAF
2
DA. The cells were incubated at 37℃ in a humidified incubator with 5%
257
CO
2
for 30 minutes. The media containing dye was removed and the cells were washed with
258
1XPBS and the cells were acquired immediately for flow-cytometry (BD FACS Verse) (Ex-
259
491nm/Em- 513nm).
260
Intracellular Glutathione determination
261
The intracellular reduced Glutathione (GSH) concentration was determined by modification of a
262
pubmished protocol [15]. Briefly, a standard curve with known concentration of GSH (Sigma) was
263
prepared. Reaction mixture for each contained 600µL of phosphate buffer (0.1M, pH7), 40µL of
264
0.4% w/v 5,5-dithiobis(2-nitrobenzoic acid) (DTNB, from Sigma), 100 µL of the standard
265
solutions of GSH (0mM-1mM range) and autoclaved MilliQ water to make up the volume to
266
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint

1000µL.The mixture was incubated at room temperature for 5 minutes and absorbance was
267
measured at 412nm using tecan plate reader. The bacterial strains were subcultured in fresh LB
268
media and grown till OD 0.1 (exponential phase) and washed with buffer (Tris, Sodium chloride
269
and EDTA) and lysed using sonication. The supernatant after sonication was used as the sample
270
for GSH detection. As previously described for standard curve. From the standard curve the
271
intracellular concentrations were interpolated.
272
In silico analysis
273
The in silico protein structure determination was performed using the SWISS-MODEL software
274
(https://swissmodel.expasy.org/), where we supplied the protein sequence of GspSA of
275
Salmonella Typhimurium from UniProt. We analysed the model with highest sequence identity
276
and maximum coverage with the Gsp from E.coli. The structure depicts a Homo-dimer with a
277
GMQE of 0.93 and QMEANisCo Global of 0.88 ± 0.05.
278
In vivo animal experiment
279
5-6weeks old C57BL/6 mice were infected by orally gavaging 10
7
CFU of STM WT, STM
280
ΔpotCD, STM ΔspeED, STM ΔpotCDΔspeED, STM Δgsp and STM ΔkatG. To study the
281
colonisation in organs, the intestine (Peyer’s patches), MLN, spleen and liver were isolated
282
aseptically, 5 days post-infection, and the CFU was enumerated on differential and selective SS
283
agar by serial dilution. For intraperitoneal infection 5-6weeks old C57BL/6 mice were infected
284
by intraperitoneally injecting 10
3
CFU of STM WT, STM ΔpotCD, STM ΔspeED, STM
285
ΔpotCDΔspeED, STM Δgsp and STM ΔkatG. To study the colonisation in organs, spleen and liver
286
were isolated aseptically 3 days post-infection. Blood was isolated by heart puncture 3 days post-
287
infection. The CFU was enumerated on differential and selective SS agar by serial dilution. Organs
288
were stored in 3.5%PFA before histopathogical sample preparation.
289
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
For inhibitor treatment, 5-6weeks old C57BL/6 mice were infected by orally gavaging 10
7
CFU
290
of STM WT. The inhibitor DFMO (Sigma D193) was intraperitoneally injected every alternate
291
day from day 1 at two doses 2mg/kg and 1mg/kg of body weight of mice. To study the colonisation
292
in organs, the intestine (Peyer’s patches), MLN, spleen and liver were isolated aseptically, 5 days
293
post-infection, and the CFU was enumerated on differential and selective SS agar by serial dilution.
294
For survival assay of mice 5-6weeks old C57BL/6 mice were infected by orally gavaging 10
8
CFU
295
of STM WT. The inhibitor DFMO was intraperitoneally injected every alternate day from day 1 at
296
two doses 2mg/kg and 1mg/kg of body weight of mice. The survival was monitored till 10 days.
297
Likewise, organs were stored in 3.5% PFA before histopathogical sample preparation.
298
Mass Spectrometry to determine intracellular GS-sp levels
299
The bacterial strains were grown in LB media with added 1mM hydrogen peroxide until the log
300
phase of growth. The cells were centrifuged to remove the media, and the cells were resuspended
301
in the lysis buffer (Sodium chloride, Tris, EDTA, 10% protease inhibitor cocktail) after washing
302
with 1XPBS. The cells were lysed using sonication and centrifuged at 4°C to collect the cell lysate.
303
Protein was precipitated using ice-cold acetone (Sigma, MS grade), 4 times the volume of the cell
304
lysate and by incubating at -20°C overnight. The precipitated proteins were removed by
305
centrifugation and the supernatatnt was used for analysis. Samples were analysed by ESI MS Q-
306
TOF, impact HD ( Bruker Daltonics Germany) connected to Agilent HPLC 1260. Samples were
307
passed through Agilent C18, 4.6X150mm column. Mobile phase used was water and Acetonitrile
308
with 0.1% formic acid. Linear gradient was used with flow rate 0.2ml/min. Data was analysed
309
using Bruker Daltonics software Data analysis 4.1. Mass of GS-sp (Glutathionyl spermidine) is
310
434g/mol, and (GS-sp)
2
(oxidized form, Di-glutathionyl spermidine) is 866g/mol.
311
312
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Statistical Analysis
313
Statistical analyses were performed with GraphPad Prism software. The Student’s t-test
314
(parametric, two-tailed, unpaired) was performed as indicated. For animal experiments Non-
315
parametric Mann-whitney (two-tailed) test was performed. The results are expressed as mean ±
316
SD or mean ± SEM. Group sizes, experiment number, and p values for each experiment are
317
described in figure legends.
318
Results
319
Loss of spermidine transporter and biosynthesis genes in Salmonella Typhimurium
320
compromises its ability to be phagocytosed by the macrophages
321
The pathoadaptation in Salmonella involves multiple players, which counteracts the stressful
322
condition it encounters in the host macrophages. Polyamines being a group of well-studied stress
323
response molecules, we were interested in determining the expression of the spermidine transporter
324
and biosynthesis genes in Salmonella Typhimurium. Our previous study shows that Salmonella
325
upregulates the spermidine transporter genes (potA, potB, potC and potD) and the biosynthesis
326
genes (speE and speD) during the log phase of growth in vitro [16]. Here we assessed the mRNA
327
levels of potA, potB, potC and potD in STM WT upon infection into the RAW264.7 macrophage
328
cell line. We noted that all the genes showed a 1.5-2 fold upregulation post 6 hours of infection
329
into macrophages till 16 hours (Fig 1 A). Further, our results showed that the spermidine
330
biosynthesis enzymes speE and speD were upregulated 1.5 to 2 folds post 6 hours to 16 hours post-
331
infection into macrophages (Fig 1 B). These results indicate that Salmonella Typhimurium
332
enhances its intracellular spermidine biosynthesis and imports from the extracellular milieu. It
333
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
might be a strategy of the pathogen to increase the intracellular pool of stress response molecules
334
like the polyamine spermidine, as it encounters the hostile environment of host macrophages.
335
We then determined how the spermidine transporter and biosynthesis mutants (STM ∆potCD, STM
336
∆speED and STM ∆potCD∆speED) behave upon infection into RAW264.7 macrophages. We
337
infected STM WT, STM ∆potCD, STM ∆speED and STM ∆potCD∆speED into RAW264.7 cells
338
and observed that STM ∆potCD, STM ∆speED and STM ∆potCD∆speED showed reduced
339
phagocytosis by the macrophages compared to the wild type (Fig 1 C). Interestingly, STM
340
∆potCD, STM ∆speED and STM ∆potCD∆speED showed a compromised ability to proliferate
341
intracellularly in the RAW264.7 cells further validating a previous study [12](Fig S1 A). To further
342
validate our results, we assessed the behaviour of the spermidine transporter and biosynthesis gene
343
mutants in primary macrophages isolated from the peritoneal lavage of C57BL/6 mice. Likewise,
344
the mutants showed attenuated proliferation and uptake by phagocytosis into the peritoneal
345
macrophages (Fig 1 D and S1 B). Collectively these results suggest that spermidine is a critical
346
molecule in Salmonella Typhimurium to infect and survive in macrophages. To further investigate
347
the reason behind the reduced ability to be taken up by macrophages upon loss of spermidine
348
biosynthesis and transport, we treated STM WT, STM ∆potCD, STM ∆speED and STM
349
∆potCD∆speED with mouse complement-sera. Mouse complement-sera acts as an opsonin and
350
thus potentiates the interaction of the bacteria with the macrophages. Upon pre-treatment with
351
mouse complement-sera, we noted a rescue in the reduced uptake of the mutants by peritoneal
352
macrophages isolated from C57BL/6 mice (Fig 1 E). A study on Salmonella Typhimurium
353
revealed that the aflagellate and non-motile Salmonella collide less frequently with macrophages
354
and gets merest time to maintain contact with the macrophages, thereby showing decreased
355
phagocytosis [17]. Our group previously showed that loss of spermidine production and import in
356
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Salmonella Typhimurium results in the loss of flagella formation on the bacterial cell surface [16].
357
Thus, we determined the percentage phagocytosis for a flagellin-deficient strain of Salmonella
358
Typhimurium, STM ∆fliC and observed that STM ∆potCD, STM ∆speED, STM ∆potCD∆speE
359
and STM ∆fliC exhibited a significantly decreased ability to be taken up by the RAW264.7
360
macrophages (Fig 1 F). Furthermore, we observed a rescue of the attenuated percentage
361
phagocytosis and fold proliferation of only STM ∆speED upon supplementation of spermidine
362
(100µM) during the growth of bacteria prior to infection (Fig 1 G and S1C). We generated single
363
gene mutants for abrogating the spermidine transport (STM ∆potA) and spermidine biosynthesis
364
(STM ∆speE) function and further complemented the genes through a vector (STM ∆potA:potA
365
and STM ∆speE:speE ), we observed a recovery of the fold proliferation and percentage
366
phagocytosis nearly to STM WT in the complemented strains (Fig S1 D-E) . Hence, the plausible
367
explanation is that the reduced ability to form flagella in the spermidine mutants causes less
368
frequent interaction with the macrophages and provides minimal contact time for infection, leading
369
to reduced phagocytosis by the macrophages.
370
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Figure 1: Loss of spermidine transporter and biosynthesis genes in Salmonella Typhimurium
372
compromises its ability to be phagocytosed by the macrophages
373
A.The mRNA expression of spermidine transporter genes potA, potB, potC and potD in STM WT
374
upon infection into RAW264.7 cells, B. The mRNA expression of spermidine biosynthesis genes
375
speE and speD in STM WT upon infection into RAW264.7 cells, C. The percentage phagocytosis
376
of the spermidine mutants in RAW264.7 cells, D. The percentage phagocytosis in primary
377
macrophages isolated from peritoneal lavage of C57BL/6 mice, E. The percentage phagocytosis
378
upon pre treatment with mouse-complement sera which act as an opsonin, F. The percentage
379
phagocytosis in RAW264.7 cells with flagellin mutant (STM ∆fliC), G. The percentage
380
phagocytosis in RAW264.7 cells with the spermidine mutants grown in media supplemented with
381
100µM spermidine (SPD). Student’s t-test was used to analyze the data; p values ****<0.0001,
382
***<0.001, **<0.01, *<0.05. Two-way Anova was used to analyze the grouped data; p values
383
****<0.0001, ***<0.001, **<0.01, *<0.05.
384
385
Spermidine provides stress resistance in Salmonella Typhimurium by regulation of the
386
expression of numerous antioxidative enzymes
387
The loss of spermidine transport and biosynthesis function in Salmonella Typhimurium renders it
388
incapable of proliferation and survival in macrophages. In the host macrophages, the bacteria
389
encounter numerous threats, of which the foremost is the rapid oxidative burst mediated by the
390
NOX2. The reactive oxygen species superoxide radical can easily diffuse through the bacterial
391
membrane and pose a major threat to the pathogen. ROS acts on multiple molecules such as nucleic
392
acids, proteins and lipids, thus damaging the cell membranes, DNA and proteins within the bacteria
393
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
[18]. Spermidine has been linked to stress response against oxidative stress and protects bacteria
394
in E. coli and Pseudomonas aeruginosa. Thus, we were intrigued to understand the role of
395
spermidine in antioxidative response in Salmonella Typhimurium. We examined the survival of
396
STM WT, STM ∆potCD, STM ∆speED, STM ∆potCD∆speE upon exposure to the oxidative agent
397
hydrogen peroxide in vitro. We noticed that at high concentrations of hydrogen peroxide, 5mM
398
and 10mM, the STM ∆potCD, STM ∆speED and STM ∆potCD∆speE showed significantly lesser
399
survival than STM WT(Fig 2 A). Moreover, the complemented strains for spermidine transport
400
and synthesis mutants showed a rescue in attenuated survival in the presence of high
401
concentrations of hydrogen peroxide in vitro (Fig S2 A). We further determined the expression of
402
potA, potB, potC, potD, speE and speD in STM WT upon exposure to 1mM hydrogen peroxide.
403
There was a 2-3 fold upregulation in the mRNA expression of the transporter genes over the
404
untreated during the early log phase of growth (6 hours) and the late log phase of growth (12 hours)
405
in vitro (Fig S2 B-E). Similarly, the biosynthesis genes speE and speD were 4-6 fold upregulated
406
in their corresponding mRNA expressions during their early log phase of growth (6 hours) and the
407
late log phase of growth (12 hours) in vitro (Fig S2 F and G). Our results show that Salmonella
408
Typhimurium upregulates spermidine transport and biosynthesis upon oxidative stress suggesting
409
that spermidine mounts a protective function in such a stressful condition to aid bacterial survival.
410
Bacteria sense the environmental changes and cues to respond and adapt to the altered
411
environment. They use the two-component systems, transcriptional activators and repressors to
412
alter gene expression in response to a stimulus [19]. Polyamines in E. coli regulate multiple genes
413
at the transcription and translation together, referred to as the “Polyamine modulon”. These involve
414
the numerous mRNAs, tRNAs, sigma factors, translational factors and two-component systems
415
during the bacterial growth as well as in stress conditions [20-22]. Salmonella harbors multiple
416
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
antioxidative enzymes to detoxify the ROS intracellularly [23]. Our study so far show that STM
417
∆potCD, STM ∆speED and STM ∆potCD∆speE is attenuated in survival under in vitro oxidative
418
stress than STM WT. To gain mechanistic insight into the attenuated survival of Salmonella
419
Typhimurium we determined the mRNA expression of the critical transcription factor rpoS, which
420
activates the expression of the catalase enzymes (katG and katE) in order to detoxify hydrogen
421
peroxide to water in the bacteria enzymatically [24]. In both STM ∆potCD and STM ∆speED the
422
mRNA expression of rpoS is significantly down regulated from 6 hours post infection in
423
RAW264.7 cells (Fig 2 B and C, S3 A). Further, its downstream target katG was also down
424
regulated in STM ∆potCD and STM ∆speED from 6 hours post infection into RAW264.7 cells
425
(Fig 2 D and E, S3 B). We further assessed the mRNA expression of the transcription factor soxR,
426
which regulates the expression of superoxide dismutases (sodA and sodB) [25]. Upon infection in
427
RAW264.7 cells, expression of soxR was significantly downregulated in STM ∆potCD and STM
428
∆speED with respect to STM WT (Fig 2 F and G, S3 C). Superoxide dismutases act on superoxide
429
radicals, the potent ROS encountered in macrophages, converting to hydrogen peroxide. The
430
mRNA expression of both sodA and sodB were likewise downregulated in STM ∆potCD and STM
431
∆speED upon infection into RAW264.7 macrophages (Fig 2 H-K, S3 D and E). A major
432
antioxidant in most living organisms is glutathione (GSH), which directly acts as a quencher of
433
ROS [26]. GSH is synthesized by Glutathione synthase (GshA) which in turn is regulated by EmrR
434
transcription factor. We observed that the mRNA expression of emrR is downregulated in STM
435
∆potCD and STM ∆speED upon infection into RAW264.7 macrophages (Fig 2 L and M, S3 F).
436
Similarly, gshA transcript expression is downregulated as well (Fig 2 N and O, S3 G). To further
437
validate the down regulation of the glutathione synthesis arm in spermidine transport and
438
biosynthesis mutants, we determined the intracellular GSH levels and noted the in STM ∆potCD,
439
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
STM ∆speED and STM ∆potCD∆speED the levels were significantly less (Fig S3H and I). Taken
440
together these results indicate that spermidine regulates the transcription of multiple transcription
441
factors involved in oxidative stress response in Salmonella Typhimurium. Importantly, we found
442
a mechanism of oxidative stress resistance in Salmonella Typhimurium regulated by the
443
spermidine, potentiating the survival of the bacteria.
444
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Figure 2: Spermidine provides stress resistance in Salmonella Typhimurium by regulation of
446
the expression of numerous antioxidative enzymes
447
A.The in vitro hydrogen peroxide sensitivity assay with the spermidine transport and biosunthesis
448
mutants, B. The mRNA expression of stress responsive transcription factor rpoS in STM ∆potCD
449
upon infection into RAW264.7 cells, C. The mRNA expression of stress responsive transcription
450
factor rpoS in STM ∆speED upon infection into RAW264.7 cells, D. The mRNA expression of
451
katG in STM ∆potCD upon infection into RAW264.7 cells, E. The mRNA expression of katG in
452
STM ∆speED upon infection into RAW264.7 cells, F. The mRNA expression of oxidative stress
453
responsive transcription factor soxR in STM ∆potCD upon infection into RAW264.7 cells, G. The
454
mRNA expression of oxidative stress responsive transcription factor soxR in STM ∆speED upon
455
infection into RAW264.7 cells, H. The mRNA expression of sodA in STM ∆potCD upon infection
456
into RAW264.7 cells, I. The mRNA expression of sodA in STM ∆speED upon infection into
457
RAW264.7 cells, J. The mRNA expression of sodB in STM ∆potCD upon infection into
458
RAW264.7 cells, K. The mRNA expression of sodB in STM ∆speED upon infection into
459
RAW264.7 cells, L. The mRNA expression of glutathione synthetase specific transcription factor
460
emrR in STM ∆potCD upon infection into RAW264.7 cells, M. The mRNA expression of
461
glutathione synthetase specific transcription factor emrR in STM ∆speED upon infection into
462
RAW264.7 cells, N. The mRNA expression of gshA in STM ∆potCD upon infection into
463
RAW264.7 cells, O. The mRNA expression of gshA in STM ∆speED upon infection into
464
RAW264.7 cells.
465
466
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
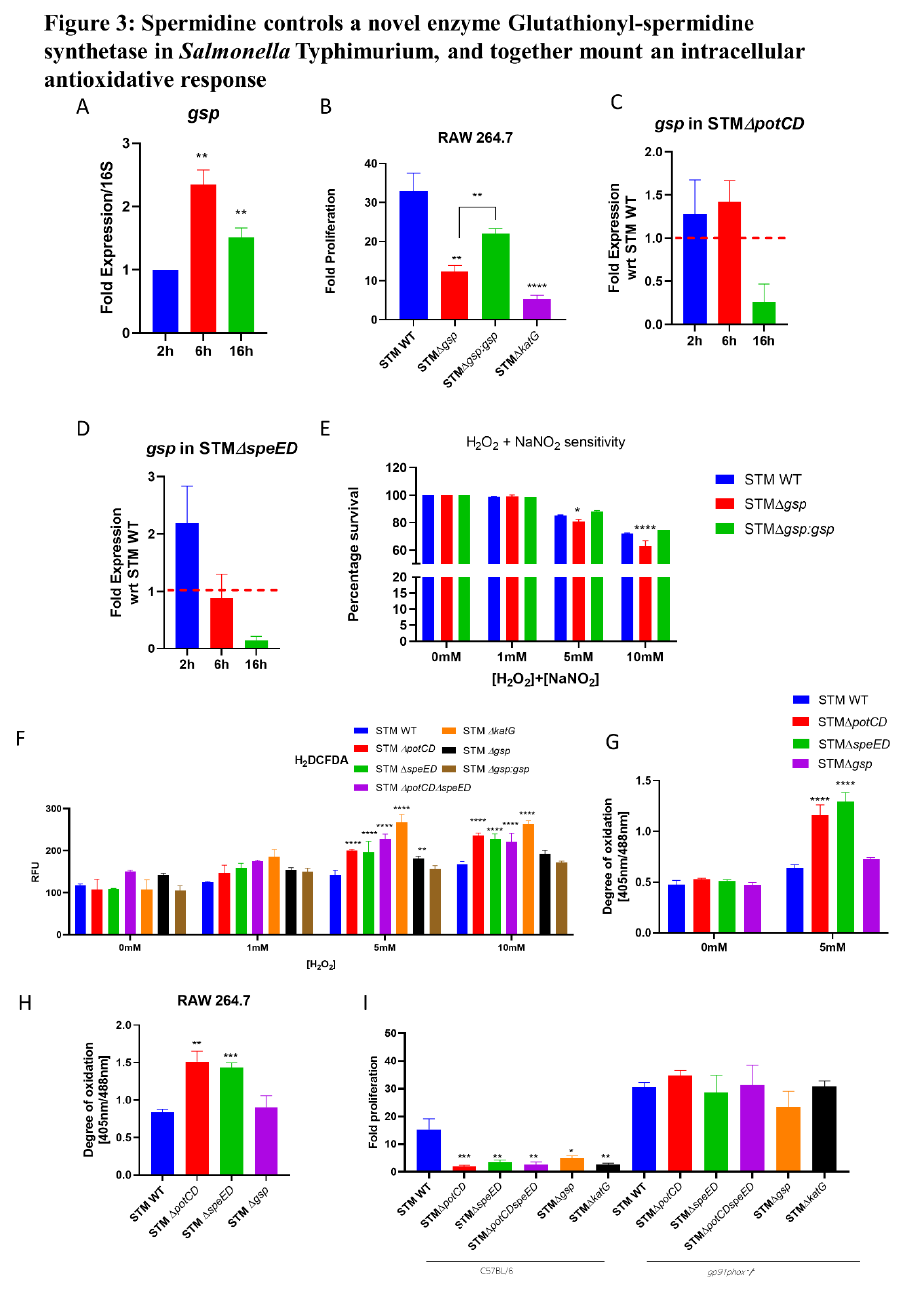
Spermidine controls a novel enzyme Glutathionyl-spermidine synthetase in Salmonella
467
Typhimurium, and together mount an intracellular antioxidative response
468
The spermidine synthesized from putrescine has two fates. It is either acetylated by the enzyme
469
SpeG or it is covalently conjugated to GSH to form Glutathionyl spermidine (GS-sp) catalysed by
470
the enzyme Glutathionyl-spermidine synthetase (Gss). Tabor (1974) first discovered the existence
471
of this enzyme in E. coli [27]. In E. coli GS-sp is generated at a higher level in the stationary phase
472
and very less in the late exponential phase. It also interacts and modifies the thiol containing
473
proteins under high H
2
O
2-
containing media leading to the formation of Gsp-thiolated proteins (PS-
474
Gsp). Certain in vitro experiments with dehydro-ascorbate suggested that GS-sp might have higher
475
antioxidant properties than GSH [28] and may be more effective in protecting against DNA
476
damage by free radicals [29]. However, in E. coli Gss could not be linked to its pathogenicity [30].
477
Among Enterobacteriaceae, Salmonella was found to possess this unique enzyme. Our in silico
478
analysis suggested that the enzyme in Salmonella Typhimurium (GspSA, encoded by gsp) has 90%
479
identity with E. coli gss and the SWISS MODEL predicts it to be a homo-dimeric protein (Fig S4
480
A-C). Also, the spermidine synthesised by SpeE in Salmonella Typhimurium is directly fed into
481
the pathway to synthesise GS-sp. We thus, investigated biological role of this novel enzyme in
482
Salmonella Typhimurium. We noted that the mRNA expression of gsp is significantly upregulated
483
at the late-log phase of growth of STM WT in LB media in the presence of H
2
O
2
(Fig S4 D). Upon
484
infection into RAW264.7 cells, STM WT upregulates the mRNA expression of gsp at 6 hours and
485
16 hours post-infection into RAW264.7 cells (Fig 3 A). Further, the Salmonella Typhimurium
486
mutant of gsp (STM ∆gsp) showed attenuated proliferation in RAW264.7 cells similar to STM
487
∆katG, which has reduced capability to detoxify ROS (Fig 3 B). Thus, our results suggest that gsp
488
is important in Salmonella Typhimurium to survive and cope with the oxidative stress and hostile
489
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
environment of macrophages. Interestingly, the mRNA expression of gsp was found to be
490
downregulated in both STM ∆potCD and STM ∆speED at 16 hours post-infection into RAW264.7
491
macrophages (Fig 3 C and D). These findings show that spermidine maintains its intracellular
492
levels by regulating the flux of spermidine into other pathways, such as the GspSA pathway. Also,
493
it further potentiates the ability of Salmonella Typhimurium to mount an antioxidative response by
494
regulating gsp expression.
495
GspSA enzyme in Trypanosomes was named Trypanothione synthetase, while the conjugate is
496
called Trypanothione (TSH). In trypanosomes TSH is essential, and these organisms entirely rely
497
on TSH and possess no GSH reductase. Also, they have evolved to use TSH reductase instead of
498
GSH reductase, Glutaredoxins or Thioredoxins [31]. In E. coli GS-sp forms bonds with the
499
Cysteine thiol groups of numerous proteins and protect them from oxidation under oxidative stress.
500
Cysteine thiol groups are highly prone to attack by ROS and get oxidized to sulphinic and
501
sulphonic acids. In Salmonella Typhimurium we observe that loss of gsp results in attenuated
502
proliferation and survival in macrophages. Thus, we investigated the survival of STM ∆gsp upon
503
exposure to hydrogen peroxide in vitro. STM ∆gsp exhibited reduced survival in the presence of
504
high 5mM and 10mM concentrations of H
2
O
2
(Fig S4 E). Likewise, upon exposure to agents of
505
oxidative stress and nitrosative stress H
2
O
2
and NaNO
2
together, we observe that STM ∆gsp
506
exhibited reduced survival at higher concentrations such as 5mM and 10mM (Fig 4 E). Thus, gsp
507
is critical in Salmonella Typhimurium to shield the bacteria from the action of ROS and RNS. As
508
we observed that spermidine transporter and biosynthesis mutants and gsp mutant of Salmonella
509
Typhimurium are compromised in their survival under oxidative stress and in macrophages thus,
510
we were interested to assess the intracellular ROS detoxification abilities of the strains. We
511
determined the intracellular ROS in STM WT, STM ∆potCD, STM ∆speED, STM ∆potCD∆speE,
512
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
STM ∆gsp and STM ∆katG. Interestingly, STM ∆potCD, STM ∆speED, STM ∆potCD∆speE,
513
STM ∆gsp and STM ∆katG showed significantly higher intracellular ROS levels when they were
514
exposed to 5mM and 10mM H
2
O
2
(Fig 3 F). Further the complemented strains for spermidine
515
transport and synthesis mutants showed a reduced intracellular ROS in the presence of high
516
concentrations of hydrogen peroxide in vitro (Fig S4 F). These results suggest that lower
517
intracellular levels of spermidine correlate to higher intracellular ROS levels in Salmonella
518
Typhimurium. To validate our observed results, we utilised a genetically engineered tool to sense
519
the redox status of the bacterial cytosol, roGFP2 is a genetically modified form of GFP, and we
520
use pQE60-Grx1-roGFP2 plasmid. The glutaredoxin (Grx1) fused to the roGFP2, reversibly
521
transfers electrons between the cytosolic pool of GSH/GSSG and the thiol group of roGFP2, and
522
the ratio of fluorescence ratio at 408nm and 488nm determine the redox status of bacterial
523
cytoplasm [14]. We observed that STM ∆potCD, STM ∆speED harbouring the pQE60-Grx1-
524
roGFP2 showed higher ratio of 405nm/488nm compared to STM WT in the presence of 5mM
525
H
2
O
2
in vitro and also upon infection into RAW 264.7 macrophages (Fig 3 G and H). However,
526
STM ∆gsp did not show a significantly higher ratio of 405nm/488nm. Moreover upon
527
supplementation of the growth media with 100 µM spermidine, only in STM ∆speED we observed
528
a lower intracellular ROS and lesser 405nm/488nm in higher concentration of H
2
O
2
(Fig S4 G
529
and H). Thus, our results indicate that spermidine is critical in mounting an antioxidative response
530
to detoxify the intracellular ROS, by regulating multiple antioxidant genes in Salmonella
531
Typhimurium. To validate our observed results we determined the intracellular levels of
532
glutathionyl-spermidine in STM WT, STM ∆potCD, STM ∆speED and STM ∆gsp using mass
533
spectrometry. Our study qualitatively shows that the synthesis of GS-sp and (GS-sp)
2
(oxidized
534
form, di-glutathionylspermidine) in STM WT upon exposure to 1mM hydrogen peroxide, which
535
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
is absent in the spermidine mutants and STM ∆gsp (Fig S5 A and B). We further determined the
536
presence of PS-Gsp and show that STM WT shows modifications of proteins by GS-sp (detected
537
by anti-spermidine antibody), which is reduced upon treatment with beta-mercaptoethanol. The
538
spermidine mutants show very less modification, while it is almost negligible in STM ∆gsp (Fig
539
S5 C-E).
540
We observe that STM ∆potCD, STM ∆speED show a higher intracellular ROS than STM ∆gsp.
541
Thus to understand does STM ∆gsp phenocopy STM ∆potCD, STM ∆speED, we generated double
542
mutants STM ∆gsp∆potCD and STM ∆gsp∆speED. We find that, the double mutants show a
543
similar reduced fold proliferation and percentage phagocytosis in RAW 264.7 cells (Fig S6 A-B).
544
However, they show an enhanced intracellular ROS upon exposure to hydrogen peroxide (Fig S6
545
C). Upon co-infection of the double mutants with STM ∆gsp in C57BL/6 mice, we see that STM
546
∆gsp out competes the double mutants in colonizing the liver (Fig S6 D-E). To further dissect the
547
role of spermidine in protection of Salmonella Typhimurium from oxidative stress, we infected
548
STM WT, STM ∆potCD, STM ∆speED, STM ∆potCD∆speE, STM ∆gsp and STM ∆katG in
549
primary macrophages isolated from the peritoneal lavage of gp91phox -/- mice. Gp91Phox is the
550
major subunit of the NOX2 complex, that aids in the catalysis of oxygen to superoxide radical.
551
Interestingly we observe a rescue in the attenuated fold proliferation of STM ∆potCD, STM
552
∆speED, STM ∆potCD∆speE, STM ∆gsp and STM ∆katG in peritoneal macrophages isolated
553
from gp91phox -/- mice (Fig 3 I). Our results demonstrate the vital role of spermidine in oxidative
554
stress resistance in Salmonella Typhimurium.
555
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Figure 3: Spermidine controls a novel enzyme Glutathionyl-spermidine synthetase in
557
Salmonella Typhimurium, and together mount an intracellular antioxidative response
558
A.The mRNA expression of gsp in STM WT upon infection into RAW264.7 cells, B The fold
559
proliferation of STM WT, STM ∆gsp and STM ∆gsp:gsp in RAW264.7 cells, C. The mRNA
560
expression of gsp in STM ∆potCD upon infection into RAW264.7 cells, D. The mRNA expression
561
of gsp in STM ∆speED upon infection into RAW264.7 cells, E The in vitro Hydrogen peroxide
562
and nitic oxide sensitivity assay, F. Intracellular reactive oxygen species determination using the
563
cell permeable H
2
DCFDA dye, G. The intracellular degree of oxidation in spermidine and gsp
564
mutants using pQE60-grx1-roGFP2 construct in vitro, H. The intracellular degree of oxidation
565
using pQE60-grx1-roGFP2 construct in spermidine and gsp mutants upon infection into RAW
566
264.7 cells , I. The fold proliferation in primary macrophages isolated from wild type C57Bl/6
567
mice and gp91phox-/- mice, Student’s t-test was used to analyze the data; p values ****<0.0001,
568
***<0.001, **<0.01, *<0.05. Two-way Anova was used to analyze the grouped data; p values
569
****<0.0001, ***<0.001, **<0.01, *<0.05.
570
571
Spermidine is critical for Salmonella Typhimurium to colonise the primary and secondary
572
sites of infection in mice
573
Salmonella infects the host and breaching the epithelial cells at the Peyer’s patches in the distal
574
ileum, it disseminates to the secondary sites of infection namely the Mesentric Lymph node
575
(MLN), spleen and liver. From the basolateral surface of the epithelial cells at the lamina propria,
576
Salmonella is taken by the macrophages and polymorphonuclear cells (PMN). We observed that
577
the spermidine transporter and biosynthesis mutants show attenuated survival in macrophages and
578
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
under oxidative stress in vitro. We were intrigued to study the behaviour of the mutants during in
579
vivo colonisation in the mouse model of Salmonella infection. We infected C57BL/6 mice by orally
580
gavaging with STM WT, STM ∆potCD, STM ∆speED, STM ∆potCD∆speE, STM ∆gsp and STM
581
∆katG at a CFU of 10
7
per mice. We noted that STM ∆potCD, STM ∆speED, STM ∆potCD∆speE,
582
STM ∆gsp and STM ∆katG showed a significantly lower organ burden in Peyer’s patches, MLN,
583
spleen and liver upon oral gavage (Fig 4 A-E). Our previous study showed that the spermidine
584
transporter and biosynthesis mutants exhibit a poor invasiveness into IECs, which can explain our
585
in vivo colonisation upon oral gavage [16]. Oral gavage mimics the physiological route of
586
Salmonella infection into its host. Hence, it requires to be able to breach the intestinal barrier
587
successfully. Incapability to invade the IECs in STM ∆potCD, STM ∆speED, STM ∆potCD∆speE
588
and STM ∆gsp explains the diminished colonisation in the organs. To dissect the role of spermidine
589
in in vivo colonisation we infected C57Bl/6 mice intraperitoneally, by bypassing the entry by
590
breaching epithelial barrier. We observed that upon infecting intraperitoneally, STM ∆potCD, STM
591
∆speED, STM ∆potCD∆speE, STM ∆gsp and STM ∆katG exhibited reduced colonisation in
592
spleen and liver and less dissemination in blood compared to STM WT (Fig 4 F-I). Also, the
593
histopathological sections show significantly less liver tissue damage with STM ∆potCD, STM
594
∆speED, STM ∆potCD∆speE, STM ∆gsp and STM ∆katG , which is validated by disease scoring
595
of the same(Fig 4 J and K). Our results thus show that spermidine aids in the in vivo pathogenesis
596
and virulence of Salmonella Typhimurium.
597
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Figure 4: Spermidine is critical for Salmonella Typhimurium to colonise the primary and
599
secondary sites of infection in mice
600
A.The experimental protocol for organ burden in C57BL/6 mice by orally gavaging 10
7
CFU per
601
mice, B. The organ burden post 5 days of oral gavage in intestine, C. in Mesentric lymph node
602
(MLN), D. in spleen, E. in liver, F. The experimental protocol for organ burden in C57BL/6 mice
603
upon intraperitoneal (I.P) infection with 10
3
CFU per mice, G. The organ burden 3 days post I.P
604
infection in spleen, H. in liver, I. dissemination in blood, J. The hematoxylin and eosin staining of
605
the sections of liver 3 days post I.P infection of C57Bl/6 mice, K. Disease score from the
606
histopathogical sections of liver, Here, Small necrotic areas (N), Congestion and damage to the
607
endothelial lining of the central vein (C), Congestion of the hepatic portal vein and poor hepatic
608
architecture (HPV), Inflammatory immune cells (IC). The disease score is as :0 for normal
609
pathology, 1 for mild/ minor pathology, 2 for moderate pathology, and 3 for severe pathological
610
changes. Mann Whitney test was used to analyse organ burden in mice; p values ****<0.0001,
611
***<0.001, **<0.01, *<0.05. Student’s t-test was used to analyze the data; p values ****<0.0001,
612
***<0.001, **<0.01, *<0.05.
613
614
Salmonella rewires host polyamine metabolism to potentiate its survival within host
615
macrophages
616
Most of the intracellular pathogens establish their persistence in the phagocytic cells and are often
617
found to be associated with different populations of the macrophages. Like Brucella abortus
618
preferentially resides in the Alternatively activated macrophages (AAM), where it survives and
619
replicates by exploiting the host polyamines. A research group has shown that the metabolism of
620
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
AAM is shifted to increase polyamine biosynthesis by Brucella abortus and thereby promote the
621
bacterial survival [32]. Similarly, Salmonella also resides in macrophages, a way to lead chronic
622
infections. We were interested to know whether Salmonella exploits the host polyamines and leads
623
to a rewiring of host cell metabolism. To understand the host-pathogen relationship in depth, we
624
assessed the mRNA expression of Ornithine decarboxylase 1 (mOdc, mouse Ornithine
625
decarboxylase) which catalyses the rate-determining step of polyamine biosynthesis and
626
Spermidine synthase (mSrm, mouse Spermidine synthase) that synthesises spermidine from
627
putrescine by transferring the aminopropyl group from decarboxylated-S-adenosyl methionine in
628
RAW264.7 cells upon infection with STM WT. We observed that mRNA expression of mOdc1
629
and mSrm were upregulated at 6 and 16 hours post-infection (Fig 5 A and B). Further, the mRNA
630
expression of mOdc1 and mSrm were enhanced in the spleen and liver of C57BL/6 mice 5 days
631
post oral gavage with STM WT (Fig C- F). Our results show that Salmonella Typhimurium upon
632
infection into the host enhances the expression of host polyamine biosynthesis genes. Moreover,
633
we have previously observed that the Salmonella Typhimurium that cannot import spermidine
634
cannot survive and proliferate as much as STM WT in macrophages. To further delve into the role
635
of host acquired polyamines, we knocked down Odc1 in RAW264.7 cells (Fig S6 A). Upon
636
knockdown of mOdc1, Salmonella Typhimurium showed significantly attenuated proliferation in
637
RAW264.7 cells (Fig 5 G). However, the knockdown of mOdc1 did not alter the percentage of
638
phagocytosis by the macrophages (Fig S7 C). Similarly, we knocked down Srm in RAW264.7 cells
639
and observed that knock-down of spermidine synthase in the host compromises the ability of STM
640
WT to proliferate and get phagocytosed by the macrophages (Fig S7 B, D and E).
641
The question that arises is how does Salmonella regulate the host polyamine metabolic pathways?
642
Salmonella utilizes Salmonella pathogenicity island 1 and 2 encoded effectors for its entry and
643
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
survival in the specialized host niches [33, 34]. Although SPI-1 is well studied in the initial process
644
of invasion, recent studies suggests that both SPI-1 and SPI-2 effectors are required for SCV
645
maturation and Salmonella survival in the host cells [35-37]. Thus to investigate how Salmonella
646
modulates host cell polyamine metabolism, we infected RAW264.7 cells with SPI-1 mutant (STM
647
∆invC) and a SPI-2 mutant (STM ∆ssaV), both of which lack the ability to import effectors of SPI-
648
1 and SPI-2 respectively. We observed that the mRNA expression of mOdc1 and mSrm, were
649
significantly downregulated in macrophages infected with STM ∆invC and STM ∆ssaV compared
650
to in STM WT (all normalized to Unifected) (Fig 5H and I). We further determined the
651
intracellular spermidine by immunofluorescence and found that macrophages infected with STM
652
∆invC and STM ∆ssaV showed reduced spermidine production compared to in STM WT and
653
uninfected cells (Fig 5J and K). Thus, our data suggests that Salmonella utilizes effectors from
654
SPI-1 and SPI-2 to modulate the host cell polyamine metabolic pathways.
655
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Figure 5: Salmonella rewires host polyamine metabolism to potentiate its survival within host
657
macrophages
658
A.The mRNA expression of mOdc1 (mouse ornithine decarboxylase) in RAW264.7 cells upon
659
infection with STM WT, B. The mRNA expression of mSrm (mouse spermidine synthase) in
660
RAW264.7 cells upon infection with STM WT, C. The mRNA expression of mOdc1 (mouse
661
ornithine decarboxylase) in liver of C57BL/6 mice 5 days post infection with STM WT by oral
662
gavage, D. The mRNA expression of mSrm (mouse spermidine synthase) in liver of C57BL/6 mice
663
5 days post infection with STM WT by oral gavage,E. The mRNA expression of mOdc1 (mouse
664
ornithine decarboxylase) in spleen of C57BL/6 mice 5 days post infection with STM WT by oral
665
gavage, F. The mRNA expression of mSrm (mouse spermidine synthase) in spleen of C57BL/6
666
mice 5 days post infection with STM WT by oral gavage, G. The fold proliferation of STM WT in
667
RAW264.7 cells upon transient knockdown of mOdc1, H. The percentage phagocytosis of STM
668
WT in RAW264.7 cells upon transient knockdown of mOdc1. Here SCR is Scrambled (no target
669
for knock-down), two different targeted shRNA were used for knock-down purposes, Sh1is
670
shRNA-1 for knock-down, Sh2 is shRNA-2 for knock-down, and Sh1+Sh2 indicates where both
671
the shRNAs were used to obtain the knock-down, H. The mRNA expression of mOdc1 (mouse
672
ornithine decarboxylase) in RAW264.7 cells upon infection with STM WT, STM ∆ssaV, and STM
673
∆invC, normalized to the expression in uninfected macrophages, I. The mRNA expression of mSrm
674
(mouse spermidine synthase) in RAW264.7 cells upon infection with STM WT, STM ∆ssaV, and
675
STM ∆invC, normalized to the expression in uninfected macrophages, J. Immunofluorescence
676
imaging to study spermidine in RAW 264.7 cells upon infection with STM WT, STM ∆invC, STM
677
∆ssaV, here green is Anti-mouse LAMP1(Alexa fluor 488), Red is pFPV-M-cherry expressing
678
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Salmonella strains, magenta is anti-Spermidine (Alexa fluor 647) and UI- uninfected. Student’s
679
t-test was used to analyze the data; p values ****<0.0001, ***<0.001, **<0.01, *<0.05.
680
681
The chemopreventive drug DFMO, reduces Salmonella Typhimurium burden in the host by
682
enhancing nitric oxide production
683
Polyamines are essential molecules in eukarotes, with multiple roles in differentiation,
684
proliferation and development. Many studies have shown that polyamine levels are upregulated in
685
cancer cells and elevated levels of polyamines are associated with breast cancer, neuroblastoma,
686
hepatocellular carcinoma, prostate cancer, lung cancer, colorectal cancer and lukemia [38-42].
687
D,L-α-difluoromethylornithine (DFMO) , an inhibitor of ODC was developed as a potent drug to
688
treat cancer in the year 1970 [43]. DFMO irreversibly binds to the active site of ODC and acts as
689
a suicide inhibitor, thereby reducing polyamine levels and having a cytostatic effect. As a single
690
therapeutic agent it was found to be effective only in neuroblastoma , and cinical trial for other
691
cancer type was unsatisfactory [44, 45]. However, DFMO has been successfully developed as a
692
chemopreventive drug, with FDA approval for treatment of Human African Trypanosomiasis
693
(HAT) [46, 47]. To test whether DFMO can be used as a therapeutic drug against Salmonella
694
infection, we treated RAW264.7cells with DFMO during the infection with Salmonella
695
Typhimurium, and observed a significant attenuation in the fold proliferation of STM WT (Fig 6
696
A). Studies have shown that DFMO binds to ODC to prevent further production of putrescine from
697
ornithine and also acts on Arginase1 and reduces the available pool of ornithine for polyamine
698
biosynthesis [48, 49]. This, ensures the flux of arginine to be fed into the nitric oxide synthase
699
(NOS2) pathway and leads to elevated levels of nitric oxide in the cell, which in turn negatively
700
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
acts on ODC to further block polyamine synthesis [50].We assessed the mRNA expression of
701
mNos2 upon knockdown of mOdc1 in RAW264.7 cells followed by infection with STM. We
702
observed an upregulation of mOdc1 mRNA levels at 6 hours and 16 hours post infection in
703
RAW264.7 cells (Fig 6 B). Further we determined the levels of nitric oxide using a cell permeable
704
dye DAF-2DA, and noted that upon treatment with DFMO there was higher production of nitric
705
oxide (Fig 6 C and D). Taken together our results demonstrate the role of DFMO in diminishing
706
the survival and proliferation of Salmonella Typhimurium by blocking Odc1 and enhanced
707
production of nitric oxide in murine macrophages.
708
To further address whether DFMO also act as potent anti-Salmonella drug in mice model of
709
infection, we infected C57BL/6 mice with 10
7
CFU per mice by oral gavage. Every alternate day
710
the mice were injected with DFMO in two different doses for two different cohorts (2mg/kg of
711
body weight of mice and 1mg/kg of body weight of mice). Five days post oral gavage we observed
712
that DFMO treatment of 2mg/kg body weight of mice significantly reduced the colonisation of
713
STM WT in intestine, MLN, spleen and liver than in the untreated mice (Fig 6 E-I). Further, using
714
immunofluorescence we observed DFMO significantly reduced the colonisation of STM and and
715
the levels of spermidine in mouse ileum (Fig S8 A-D). Moreover, treatment of mice with DFMO
716
at a dose of 2mg/kg body weight of mice, increased the survival of mice upon infection with STM
717
WT (Fig 6 J and K). Also, the weight reduction in mice treated with with DFMO at a dose of
718
2mg/kg body weight, was less upon infection with STM WT (Fig S8 E). Next, we evaluated the
719
tissue damage upon infection of STM in mice liver. The results show that DFMO treatment of
720
mice at a dose of 2mg/kg of body weight significantly lowered the disease score, suggesting lesser
721
tissue damage compared to untreated (Fig 6 L and M ). Thus, DFMO serves as a potential drug to
722
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
treat Salmonella infection in mice by reducing the bacterial burden and tissue damage in mice and
723
enhancing the survival of mice upon infection with STM.
724
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Figure 6: The chemopreventive drug DFMO, reduces Salmonella Typhimurium burden in
726
the host by enhancing nitric oxide production
727
A. The intracellular fold proliferation of STM WT in RAW264.7 cells upon treatment with
728
Difluoromethyl ornithine (DFMO) (5µM), quantified by Gentamicin protection assay, B. The
729
mRNA expression of mNos2(mouse Nitric oxide synthase) in RAW264.7 cells, upon transient
730
knockdown of Odc1 followed by infection with STM WT, the expression is normalized to beta-
731
actin as an internal control, C. The representative scatter plot for DAF2DA dye positive population
732
of RAW264.7 cells, upon transient knockdown of Odc1 followed by infection with STM WT, the
733
data is determined by flow-cytometry post staining the infected macrophages with the dye, D. The
734
quantification of (C), E. The experimental procedure to study the organ burden of STM WT in
735
C57BL/6 mice upon treatment with DFMO, F-I. The organ burden of STM WT in Intestine, MLN,
736
Spleen and Liver of C57BL/6 mice upon intraperitoneal treatment of DFMO (2mg/kg and 1mg/kg
737
of body weight) as mentioned in (E), J. The Experimental procedure to study the survival of
738
C57BL/6 mice upon infection with STM WT and treatment with DFMO, K.The survival of
739
C57BL/6 mice upon infection with STM WT and upon intraperitoneal treatment of DFMO
740
(2mg/kg and 1mg/kg of body weight) as mentioned in (J), L. Hematoxylin and eosin staining of
741
histopathological sections of liver upon DFMO treatment to C57BL/6 mice. Here, Small necrotic
742
areas (N), Congestion and damage to the endothelial lining of the central vein (C), Congestion of
743
the hepatic portal vein and poor hepatic architecture (HPV), Inflammatory immune cells (IC), M.
744
The disease score index for liver tissue damage upon STM WT infection in C57BL/6 mice with
745
DFMO treatment. The disease score is as :0 for normal pathology, 1 for mild/ minor pathology, 2
746
for moderate pathology, and 3 for severe pathological changes, Student’s t-test was used to analyze
747
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
the data; p values ****<0.0001, ***<0.001, **<0.01, *<0.05. Two-way Anova was used to
748
analyze the grouped data; p values ****<0.0001, ***<0.001, **<0.01, *<0.05.
749
750
Discussion
751
A significant part of the life cycle of Salmonella during its pathogenesis involves its stay inside
752
the macrophages. The Gram-negative bacteria come across multiple host environmental stress
753
conditions inside the macrophages [51]. A key mechanism by which the host macrophages try to
754
limit the invading pathogen is by the rapid oxidative burst and ROS production. ROS includes
755
superoxide radicals, hydroxyl radicals, peroxy-nitrites, peroxy-chlorides and hydrogen peroxide.
756
ROS can pass through the bacterial cell wall and act on lipids, proteins and nucleic acids by
757
oxidising them and leading to cellular damage. Salmonella is often referred to as a smart pathogen.
758
Over the years, it has evolved to possess multiple strategies to combat the host-derived stresses
759
[52, 53]. To combat the oxidative burst generated NOX2 in macrophages, Salmonella possesses
760
multiple antioxidant enzymes such as the catalases KatE and KatG, the superoxide dismutases
761
SodA and SodB, the Alkyl hydroperoxide reductase, the glutaredoxins and thioredoxins and Hrg
762
transcriptional regulator [25]. Polyamines assist in Salmonella virulence and aid in stress
763
resistance. However, the mechanism behind the role of polyamines in Salmonella stress resistance
764
and virulence remains less appreciated.
765
Our study identifies spermidine as a stress-responsive regulatory molecule in Salmonella
766
Typhimurium. We show spermidine is critical for the survival and proliferation of STM in
767
macrophages and in the presence of oxidative stress in vitro. The spermidine transporter and
768
biosynthesis mutants show significant reduced capability to be phagocytosed by the macrophages.
769
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Findings from our previous study showed that the intracellular level of spermidine is significantly
770
less in both the spermidine transport and biosynthesis mutants, which further explains the
771
attenuated proliferation of both the mutants in macrophages [16]. The previous findings also
772
showed that in absence of the transport genes in Salmonella Typhimurium the synthesis genes are
773
down regulated and vice versa. The absence of spermidine transport and biosynthesis diminishes
774
the mRNA expression of multiple arms of oxidative stress response in Salmonella. Multiple studies
775
show the role of polyamines in the regulation of transcription of multiple genes by interacting with
776
DNA in eukaryotes. They bind to the DNA and change conformation as in C-MYC transcription,
777
and in other cases, enhance DNA-protein binding affinities like for Estrogen-response elements
778
[54, 55]. Thus, in Salmonella Typhimurium rpoS, soxR and emrR fall under the “Polyamine
779
modulon”.
780
We further characterise a novel enzyme GspSA in Salmonella Typhimurium, which synthesises a
781
conjugated product of glutathione (GSH) and spermidine called GS-sp. GspSA is critical for
782
Salmonella Typhimurium to survive and proliferate in macrophages. Our study shows that the
783
absence of GspSA attenuates the survival of STM under oxidative stress conditions in vitro,
784
suggesting a vital role of GspSA in protecting STM from oxidative damage. GS-sp in E. coli carries
785
out its function by modifying protein thiol groups under oxidative stress to protect the proteins
786
from getting oxidised and damaged. In Salmonella Typhimurium, we further show that the
787
spermidine regulates the gsp expression and the subsequent production of GS-sp. We expect that
788
GS-sp generated under oxidative stress conditions and inside the macrophages, likewise interact
789
with cysteine-thiol groups in proteins to shield them from oxidative damage. MK Chattopadhyay
790
(2013) showed that this enzyme (GspSA) is specifically present in two groups of organisms namely
791
bacteria and kineto-plastids respectively and completely absent in other organisms such as human,
792
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
rats, drosophila etc., among the bacteria group in all enterobacteria showed 27-100% homology
793
and >65% identity in around 50% of the species, with E. coli [30]. Thus, the absence of GspSA in
794
eukaryotes makes it a potent drug target for treatment of Salmonella infections. Thus, spermidine
795
exerts pleiotropic effects in Salmonella, by orchestrating the multiple arms of antioxidative
796
response. It strengthens the bacteria to cope with hostile host environments. Our studies in the in
797
vivo model of Salmonella Typhimurium infection further dissect the role of spermidine in assisting
798
in the pathogenesis by enhancing its virulence properties. Moreover, it is at the nexus of multiple
799
oxidative stress response arms in Salmonella, thereby assisting in mounting an antioxidative
800
response to promote its survival in macrophages.
801
Studies over the past years have given insights into the function of polyamines in the pathogenesis
802
of multiple virulent bacteria. Many human pathogenic bacteria have developed ways to exploit,
803
interfere and manipulate the polyamine metabolism of the host to enhance their fitness within the
804
niche. As in Shigella and Vibrio spp. polyamines produced by the bacteria are critical in
805
determining virulence [56, 57]. While bacteria such as Legionella spp., which does not synthesise
806
polyamines, depend on the host-acquired polyamines for their pathogenesis [58]. Another unique
807
mechanism is observed in H. pylori which activates polyamine oxidation, thereby dysregulation
808
the innate-immune response [59]. Also, in response to H. pylori infection, the host macrophages
809
increase the arginase activity and ornithine decarboxylase activity to produce polyamines [60]. A
810
recent study showed that SARS-Cov-2 hijacks the host polyamines for its reproduction and
811
infection [61].
812
Our findings also reveal that Salmonella Typhimurium enhances the polyamine production in the
813
host upon infection using its specialized pathogenicity island encoded effectors from SPI-1 and
814
SPI-2, which might be a strategy to hijack the host polyamines for its survival. This further explains
815
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint

another reason for the reduced proliferation observed for the spermidine transport mutant in
816
macrophages. Also, the knockdown of host ornithine decarboxylase attenuates Salmonella
817
proliferation in macrophages. The upregulation of Odc1 activity manages to feed the amino acid,
818
L-arginine, into the polyamine biosynthesis and prevent nitric oxide production, which otherwise
819
would be detrimental for the pathogen. Polyamines have been a well-studied as a drug target for
820
treatment of multiple cancers. The oncogene MYC is upregulated in 70 percent of the cancer types.
821
Ornithine decarboxylase (Odc1) rate limiting enzyme of polyamine biosynthesis is
822
transcriptionally activated, directly by MYC, thereby increasing Odc1 levels in cancer [62]. RAS
823
is another essential factor in cell growth and cancer development, and it directly acts on Odc1 to
824
translationally activate Odc1 in cancer cells [63]. Polyamines biosynthesis is also associated with
825
multiple other oncogenes such as AKT1 and mTORC1 [64, 65]. We further show that upon using
826
Odc1 suicide inhibitor, DFMO, Salmonella proliferation could be diminished, and it reduces the
827
bacterial burden in mice. The FDA-approved chemopreventive drug, DFMO, for Human African
828
Trypanosomiasis treatment serves to be a potential drug to cure Salmonella infection. In the
829
devastating age of increasing antibiotic resistance, such a drug promises to effectively combat
830
deadly pathogens like Salmonella.
831
Conclusion
832
A substantial duration of the infection cycle of Salmonella involves the macrophages, which
833
present a very hostile environment to the bacteria. However, Salmonella is able to survive and
834
proliferate within host macrophages and utilizes it to disseminate to secondary sites of infection.
835
Our study identifies a novel strategy employed by Salmonella Typhimurium to counteract
836
oxidative and nitrosative stress within the host. We demonstrate that spermidine is a critical
837
regulatory molecule in Salmonella that regulates multiple antioxidative pathways along with a
838
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint

novel antioxidative enzyme (GspSA) in Salmonella, to prevent oxidative damage and assist in its
839
virulence in mice. It further rewires host polyamine metabolism in a SPI-1 and SPI-2 dependent
840
manner to prevent nitric oxide production and enhance its survival. In the era of anti-microbial
841
resistance, this study further recognizes an FDA -approved chemopreventive drug, DFMO, which
842
inhibits the host-polyamine metabolism, as a prospective antidote to treat Salmonella infection.
843
844
845
Availability of data and materials
846
All data generated and analyzed during this study, including the supplementary information files,
847
are incorporated in this article. The data is available from the corresponding author on request.
848
Ethics statement
849
All the animal experiments were approved by the Institutional Animal Ethics Committee, and the
850
Guidelines provided by National Animal Care were strictly followed during all experiments.
851
(Registration No: 435 48/1999/CPCSEA).
852
Acknowledgement
853
Prof. G. Subba Rao from MCB, IISc is duly acknowledged for providing the for knockdown
854
generation. Divisional Mass Spectrometry facility IISc and Mrs. Sunita Joshi for the MS analysis.
855
Departmental Confocal Facility. Departmental Real-Time PCR Facility, Divisional Flowcytometry
856
facility and Central Animal Facility at IISc are duly acknowledged. Mr Sumith and Ms Navya are
857
acknowledged for their help in image acquisition. Dr. Ritika Chatterjee, Mr. Amartya Mukherjee,
858
Mr. Sushovan Bhattacharyya and Ms Bhavya Joshi are also acknowledged for technical help.
859
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
Author contribution statement
860
AVN and DC conceived the study. AVN and DC designed experiments. AVN and AS performed
861
experiments. RSR analysed for the disease score of tissue samples. AVN, analyzed the data,
862
prepared the figures and wrote the manuscript draft. AVN, AS and DC reviewed and edited the
863
manuscript. DC supervised the work. All the authors read and approved the manuscript.
864
Funding
865
This work is supported by the Department of Biotechnology (DBT), Ministry of Science and
866
Technology, the Department of Science and Technology (DST), Ministry of Science and
867
Technology. DC acknowledges DAE-SRC ((DAE00195) outstanding investigator award and funds
868
and ASTRA Chair Professorship funds. The authors jointly acknowledge the DBT-IISc partnership
869
program. Infrastructure support from ICMR (Center for Advanced Study in Molecular Medicine),
870
DST (FIST), UGC-CAS (special assistance), and TATA fellowship is duly acknowledged. AVN
871
acknowledges the IISc-MHRD for financial assistance. AS acknowledges UGC for the financial
872
assistance. RSR acknowledges IISc for the financial help.
873
Conflict of Interest
874
The authors declare no conflict of interest.
875
Reference
876
877
1. Hernandez, D.J., et al., Environmental stress destabilizes microbial networks. The ISME Journal,
878
2021. 15(6): p. 1722-1734.
879
2. Gray, D.A., et al., Extreme slow growth as alternave strategy to survive deep starvaon in
880
bacteria. Nature communicaons, 2019. 10(1): p. 890.
881
3. Wineld, M.D. and E.A. Groisman, Role of nonhost environments in the lifestyles of Salmonella
882
and Escherichia coli. Applied and environmental microbiology, 2003. 69(7): p. 3687-3694.
883
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
4. Dodd, C.E., P.J. Richards, and T.G. Aldsworth, Suicide through stress: a bacterial response to sub-
884
lethal injury in the food environment. Internaonal journal of food microbiology, 2007. 120(1-2):
885
p. 46-50.
886
5. Marmion, M., et al., Survive and thrive: Control mechanisms that facilitate bacterial adaptaon
887
to survive manufacturing-related stress. Internaonal Journal of Food Microbiology, 2022. 368:
888
p. 109612.
889
6. Vazquez-Torres, A. and F.C. Fang, Salmonella evasion of the NADPH phagocyte oxidase. Microbes
890
and infecon, 2001. 3(14-15): p. 1313-1320.
891
7. Chakravory, D., I. Hansen-Wester, and M. Hensel, Salmonella pathogenicity island 2 mediates
892
protecon of intracellular Salmonella from reacve nitrogen intermediates. The Journal of
893
experimental medicine, 2002. 195(9): p. 1155-1166.
894
8. Rhee, H., E.J. Kim, and J. Lee, Physiological polyamines: simple primordial stress molecules.
895
Journal of cellular and molecular medicine, 2007. 11(4): p. 685-703.
896
9. Barbagallo, M., et al., A new piece of the Shigella pathogenicity puzzle: spermidine
897
accumulaonby silencing of the speG gene. PloS one, 2011. 6(11): p. e27226.
898
10. Johnson, L., et al., Surface-localized spermidine protects the Pseudomonas aeruginosa outer
899
membrane from anbioc treatment and oxidave stress. Journal of bacteriology, 2012. 194(4):
900
p. 813-826.
901
11. Banerji, R., P. Iyer, and S.D. Saroj, Spermidine enhances the survival of Streptococcus pyogenes
902
M3 under oxidave stress. Molecular Oral Microbiology, 2022. 37(2): p. 53-62.
903
12. Espinel, I.C., P.R. Guerra, and L. Jelsbak, Mulple roles of putrescine and spermidine in stress
904
resistance and virulence of Salmonella enterica serovar Typhimurium. Microbial pathogenesis,
905
2016. 95: p. 117-123.
906
13. Datsenko, K.A. and B.L. Wanner, One-step inacvaon of chromosomal genes in Escherichia coli
907
K-12 using PCR products. Proceedings of the Naonal Academy of Sciences, 2000. 97(12): p.
908
6640-6645.
909
14. Roy Chowdhury, A., et al., Salmonella Typhimurium outer membrane protein A (OmpA) renders
910
protecon from nitrosave stress of macrophages by maintaining the stability of bacterial outer
911
membrane. PLoS Pathogens, 2022. 18(8): p. e1010708.
912
15. Salbitani, G., C. Boone, and S. Carfagna, Determinaon of reduced and total glutathione
913
content in extremophilic microalga Galdieria phlegrea. Bio-protocol, 2017. 7(13): p. e2372-
914
e2372.
915
16. Nair, A.V., et al., Spermidine facilitates the adhesion and subsequent invasion of Salmonella
916
Typhimurium into epithelial cells via the regulaon of surface adhesive structures and the SPI-1.
917
bioRxiv, 2023: p. 2023.06. 03.543567.
918
17. Achouri, S., et al., The frequency and duraon of Salmonella–macrophage adhesion events
919
determines infecon eciency. Philosophical Transacons of the Royal Society B: Biological
920
Sciences, 2015. 370(1661): p. 20140033.
921
18. Kim, J.-S., et al., Oxidave stress acvates transcripon of Salmonella pathogenicity island-2
922
genes in macrophages. Journal of Biological Chemistry, 2022. 298(7).
923
19. Miller, J.F., J.J. Mekalanos, and S. Falkow, Coordinate regulaon and sensory transducon in the
924
control of bacterial virulence. Science, 1989. 243(4893): p. 916-922.
925
20. Chaopadhyay, M.K., et al., Polyamines smulate the level of the σ38 subunit (RpoS) of
926
Escherichia coli RNA polymerase, resulng in the inducon of the glutamate decarboxylase-
927
dependent acid response system via the gadE regulon. Journal of Biological Chemistry, 2015.
928
290(29): p. 17809-17821.
929
21. Kashiwagi, K., et al., Idencaon of the polyamine-induced protein as a periplasmic oligopepde
930
binding protein. Journal of Biological Chemistry, 1990. 265(15): p. 8387-8391.
931
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
22. Sakamoto, A., et al., Three members of polyamine modulon under oxidave stress condions:
932
two transcripon factors (SoxR and EmrR) and a glutathione synthec enzyme (GshA). PLoS One,
933
2015. 10(4): p. e0124883.
934
23. Farr, S.B. and T. Kogoma, Oxidave stress responses in Escherichia coli and Salmonella
935
typhimurium. Microbiological reviews, 1991. 55(4): p. 561-585.
936
24. Storz, G., et al., Bacterial defenses against oxidave stress. Trends in Genecs, 1990. 6: p. 363-
937
368.
938
25. Rhen, M., Salmonella and reacve oxygen species: a love-hate relaonship. Journal of innate
939
immunity, 2019. 11(3): p. 216-226.
940
26. Song, M., et al., Low-molecular-weight thiol-dependent anoxidant and annitrosave defences
941
in S almonella pathogenesis. Molecular microbiology, 2013. 87(3): p. 609-622.
942
27. Bollinger, J.M., et al., Glutathionylspermidine Metabolism in Escherichia coli.: PURIFICATION,
943
CLONING, OVERPRODUCTION, AND CHARACTERIZATION OF A BIFUNCTIONAL
944
GLUTATHIONYLSPERMIDINE SYNTHETASE/AMIDASE
∗
. Journal of Biological Chemistry, 1995.
945
270(23): p. 14031-14041.
946
28. Krauth-Siegel, R.L. and H. Lüdemann, Reducon of dehydroascorbate by trypanothione.
947
Molecular and biochemical parasitology, 1996. 80(2): p. 203-208.
948
29. Ariyanayagam, M.R. and A.H. Fairlamb, Ovothiol and trypanothione as anoxidants in
949
trypanosomads. Molecular and biochemical parasitology, 2001. 115(2): p. 189-198.
950
30. Chaopadhyay, M.K., W. Chen, and H. Tabor, Escherichia coli glutathionylspermidine
951
synthetase/amidase: phylogeny and eect on regulaon of gene expression. FEMS microbiology
952
leers, 2013. 338(2): p. 132-140.
953
31. Wyllie, S., et al., Dissecng the essenality of the bifunconal trypanothione synthetase-amidase
954
in Trypanosoma brucei using chemical and genec methods. Molecular microbiology, 2009.
955
74(3): p. 529-540.
956
32. Kerrinnes, T., et al., Ulizaon of host polyamines in alternavely acvated macrophages
957
promotes chronic infecon by Brucella abortus. Infecon and immunity, 2018. 86(3): p.
958
10.1128/iai. 00458-17.
959
33. Forest, C.G., et al., Intracellular survival of Salmonella enterica serovar Typhi in human
960
macrophages is independent of Salmonella pathogenicity island (SPI)-2. Microbiology, 2010.
961
156(12): p. 3689-3698.
962
34. Ellermeier, J.R. and J.M. Slauch, Adaptaon to the host environment: regulaon of the SPI1 type
963
III secreon system in Salmonella enterica serovar Typhimurium. Current opinion in microbiology,
964
2007. 10(1): p. 24-29.
965
35. Brawn, L.C., R.D. Hayward, and V. Koronakis, Salmonella SPI1 eector SipA persists aer entry
966
and cooperates with a SPI2 eector to regulate phagosome maturaon and intracellular
967
replicaon. Cell host & microbe, 2007. 1(1): p. 63-75.
968
36. Giacomodonato, M.N., et al., SipA, SopA, SopB, SopD and SopE2 eector proteins of Salmonella
969
enterica serovar Typhimurium are synthesized at late stages of infecon in mice. Microbiology,
970
2007. 153(4): p. 1221-1228.
971
37. Brown, N.F., et al., Salmonella pathogenicity island 2 is expressed prior to penetrang the
972
intesne. PLoS pathogens, 2005. 1(3): p. e32.
973
38. Kingsnorth, A., A.B. Lumsden, and H.M. Wallace, Polyamines in colorectal cancer. Journal of
974
Brish Surgery, 1984. 71(10): p. 791-794.
975
39. Evageliou, N.F. and M.D. Hogarty, Disrupng polyamine homeostasis as a therapeuc strategy for
976
neuroblastoma. Clinical Cancer Research, 2009. 15(19): p. 5956-5961.
977
40. Tsoi, T.-H., et al., Urinary polyamines: a pilot study on their roles as prostate cancer detecon
978
biomarkers. PLoS One, 2016. 11(9): p. e0162217.
979
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
41. Choi, Y., et al., Targeng ODC1 inhibits tumor growth through reducon of lipid metabolism in
980
human hepatocellular carcinoma. Biochemical and biophysical research communicaons, 2016.
981
478(4): p. 1674-1681.
982
42. Soda, K., The mechanisms by which polyamines accelerate tumor spread. Journal of Experimental
983
& Clinical Cancer Research, 2011. 30: p. 1-9.
984
43. Weeks, C., et al., alpha-Diuoromethylornithine, an irreversible inhibitor of ornithine
985
decarboxylase, inhibits tumor promoter-induced polyamine accumulaon and carcinogenesis in
986
mouse skin. Proceedings of the Naonal Academy of Sciences, 1982. 79(19): p. 6028-6032.
987
44. Horn, Y., P.J. Schechter, and L.J. Marton, Phase I–II clinical trial with alpha-
988
diuoromethylornithine—an inhibitor of polyamine biosynthesis. European Journal of Cancer and
989
Clinical Oncology, 1987. 23(8): p. 1103-1107.
990
45. Lewis, E.C., et al., A subset analysis of a phase II trial evaluang the use of DFMO as maintenance
991
therapy for high-risk neuroblastoma. Internaonal journal of cancer, 2020. 147(11): p. 3152-
992
3159.
993
46. Roberts, S. and B. Ullman, Parasite polyamines as pharmaceucal targets. Current
994
Pharmaceucal Design, 2017. 23(23): p. 3325-3341.
995
47. Eperon, G., et al., Treatment opons for second-stage gambiense human African
996
trypanosomiasis. Expert review of an-infecve therapy, 2014. 12(11): p. 1407-1417.
997
48. Li, H., et al., Acvies of arginase I and II are liming for endothelial cell proliferaon. American
998
Journal of Physiology-Regulatory, Integrave and Comparave Physiology, 2002. 282(1): p. R64-
999
R69.
1000
49. Bojarska, J., et al., The rst insight into the supramolecular system of D, L-α-
1001
diuoromethylornithine: A new anviral perspecve. Froners in Chemistry, 2021. 9: p. 679776.
1002
50. Bauer, P.M., et al., Nitric Oxide Inhibits Ornithine Decarboxylase viaS-Nitrosylaon of Cysteine
1003
360 in the Acve Site of the Enzyme. Journal of Biological Chemistry, 2001. 276(37): p. 34458-
1004
34464.
1005
51. HaragaA, O.M., MillerSI. Salmonellaeinterplaywith hostcells, 2008. 6(1): p. 53G66.
1006
52. Buchmeier, N.A. and F. Heron, Inducon of Salmonella stress proteins upon infecon of
1007
macrophages. Science, 1990. 248(4956): p. 730-732.
1008
53. Shen, S. and F.C. Fang, Integrated stress responses in Salmonella. Internaonal journal of food
1009
microbiology, 2012. 152(3): p. 75-81.
1010
54. Kumar, N., R. Basundra, and S. Mai, Elevated polyamines induce c-MYC overexpression by
1011
perturbing quadruplex–WC duplex equilibrium. Nucleic acids research, 2009. 37(10): p. 3321-
1012
3331.
1013
55. Thomas, T., et al., Polyamine-mediated conformaonal perturbaons in DNA alter the binding of
1014
estrogen receptor to poly (dG-m5dC). poly (dG-m5dC) and a plasmid containing the estrogen
1015
response element. The Journal of Steroid Biochemistry and Molecular Biology, 1995. 54(3-4): p.
1016
89-99.
1017
56. Prosseda, G., et al., Shedding of genes that interfere with the pathogenic lifestyle: the Shigella
1018
model. Research in microbiology, 2012. 163(6-7): p. 399-406.
1019
57. Lee, J., et al., An alternave polyamine biosynthec pathway is widespread in bacteria and
1020
essenal for biolm formaon in Vibrio cholerae. Journal of Biological Chemistry, 2009. 284(15):
1021
p. 9899-9907.
1022
58. Nasrallah, G.K., et al., Legionella pneumophila requires polyamines for opmal intracellular
1023
growth. Journal of bacteriology, 2011. 193(17): p. 4346-4360.
1024
59. Chaturvedi, R., et al., Spermine oxidase mediates the gastric cancer risk associated with
1025
Helicobacter pylori CagA. Gastroenterology, 2011. 141(5): p. 1696-1708. e2.
1026
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint
60. Chaturvedi, R., et al., Inducon of polyamine oxidase 1 by Helicobacter pylori causes macrophage
1027
apoptosis by hydrogen peroxide release and mitochondrial membrane depolarizaon. Journal of
1028
Biological Chemistry, 2004. 279(38): p. 40161-40173.
1029
61. Makhoba, X.H. and S. Makumire, The capture of host cell’s resources: The role of heat shock
1030
proteins and polyamines in SARS-COV-2 (COVID-19) pathway to viral infecon. Biomolecular
1031
Concepts, 2022. 13(1): p. 220-229.
1032
62. Bachmann, A.S. and D. Geerts, Polyamine synthesis as a target of MYC oncogenes. Journal of
1033
Biological Chemistry, 2018. 293(48): p. 18757-18769.
1034
63. Origan, S. and L.M. Shantz, Ras transformaon of RIE-1 cells acvates cap-independent
1035
translaon of ornithine decarboxylase: regulaon by the Raf/MEK/ERK and phosphadylinositol
1036
3-kinase pathways. Cancer research, 2007. 67(10): p. 4834-4842.
1037
64. Gomes, A.P., T. Schild, and J. Blenis, Adding polyamine metabolism to the mTORC1 toolkit in cell
1038
growth and cancer. Developmental cell, 2017. 42(2): p. 112-114.
1039
65. Dai, F., et al., Extracellular polyamines-induced proliferaon and migraon of cancer cells by
1040
ODC, SSAT, and Akt1-mediated pathway. An-Cancer Drugs, 2017. 28(4): p. 457-464.
1041
1042
1043
1044
1045
1046
1047
1048
1049
1050
1051
1052
1053
1054
1055
1056
1057
(which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprintthis version posted November 10, 2023. ; https://doi.org/10.1101/2023.09.29.560257doi: bioRxiv preprint